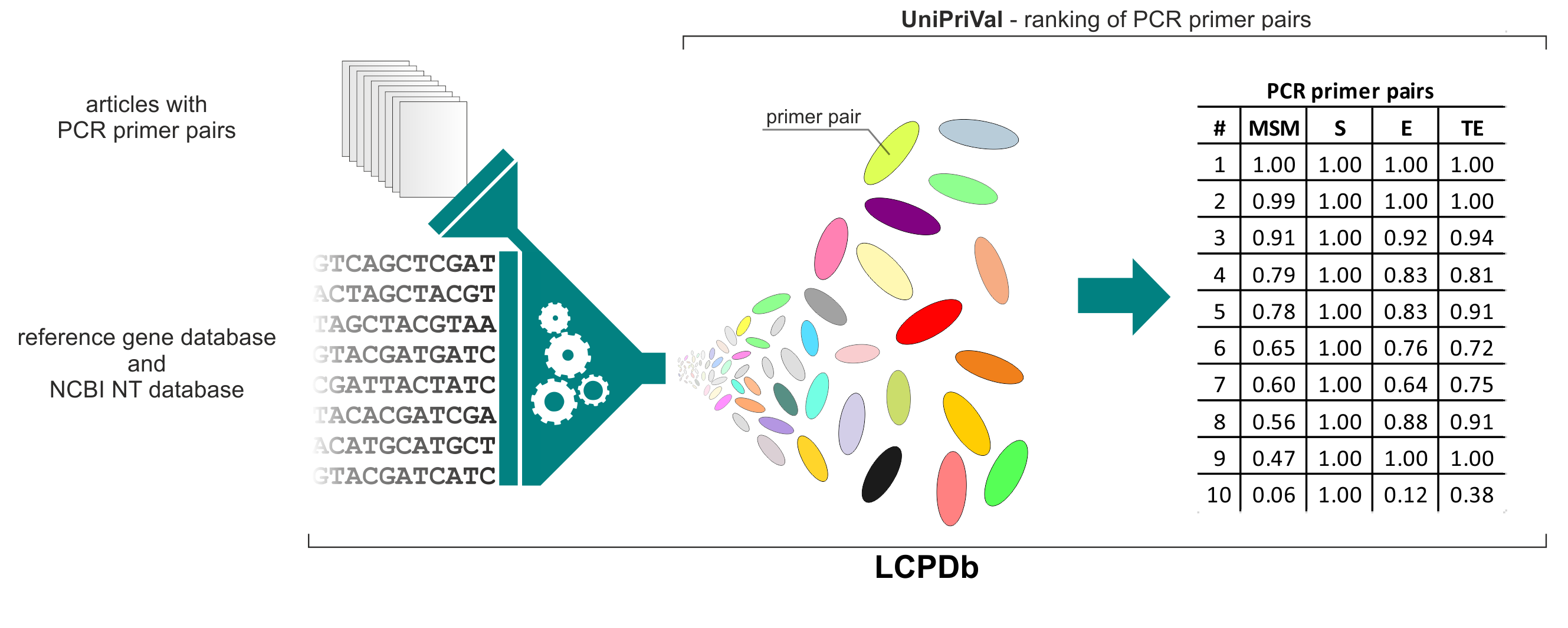
A literature-based, manually-curated database of PCR primers for the detection of antibiotic resistance genes is comprised of hundreds of PCR primer pairs designed for the amplification of various genes conferring resistance to antibiotics (LCPDb-ARG) and genes involved in metal metabolism and resistance (LCPDb-MET). Three parameters were assigned for each primer pair: specificity (S), efficacy (E) and taxonomic efficacy (TE). These parameters were evaluated using a novel bioinformatic tool – UniPriVal – that was used to validate each primer pair against various reference databases. Then, primer pairs specific for each gene were ranked based on their model success metric (MSM) value. It is important to mention, that due to the correlation between E and TE parameters, the MSM metric is biased toward primer pairs with higher E/TE values relative to S values. Despite this limitation, the internal validation system of the LCPDb application enables the quantified ranking of PCR primer pairs, which assists selection of the best primers for each application. At the end we would like to add that, although we performed thorough literature review to identify PCR primers, we are aware that the database is still incomplete and needs further development. Therefore, users are invited to directly contact (using the “Submit” webpage) the database developers to add missing primers as well as to test their own primer pairs with the UniPriVal tool.
Cite us:
LCPDb-ARG
- Gorecki, A., Decewicz, P., Dziurzynski, M., Janeczko, A., Drewniak, L., & Dziewit, L. (2019). Literature-based, manually-curated database of PCR primers for the detection of antibiotic resistance genes in various environments. Water Research, 161, 211-221. doi:10.1016/j.watres.2019.06.009 https://www.sciencedirect.com/science/article/pii/S0043135419305147
LCPDb-MET
- Dziurzynski, M., Gorecki, A., Decewicz, P., Ciuchcinski, K., Dabrowska, M., & Dziewit, L. (2022). Development of the LCPDb-MET database facilitating selection of PCR primers for the detection of metal metabolism and resistance genes in bacteria. Ecological Indicators, 145, 109606, doi:10.1016/j.ecolind.2022.109606 https://www.sciencedirect.com/science/article/pii/S1470160X22010792